🧬 Doing Biology in Julia 🧬
CAJUN
Kevin Bonham, PhD
2024-04-24
The BioJulia Organization
Brief history of BioJulia
- January 2014: First commit to
Bio.jlmonorepo- early contributors:
@dcjones,@blahah,@bicycle1885,@transgirlcodes
- early contributors:
- May 2015: Ragel introduced by
@dcjonesfor fast / accurrate parsing - April 2016: My first contribution (adding wrapper for BLAST search)
- Jan 2017:
@bicycle1885replaces Ragel with all-julia state machine generator (Automa.jl) - May 2017: Decomposition of mono repo - eg
Bio.SeqbecomesBioSequences.jl(effort driven by@transgirlcodesand@bicycle1885) - Dec 2018: Transition from
REQUIREtoProject.tomlfor julia v1.0 - Feb 2019-May 2020: Use of BioJuliaRegistry
- May 2020:
Bio.jlofficially deprecated / archived
Who we are (currently)
Project Admins
@jakni- Jakob Nybo Nissen, University of Copenhagen- Major contributions to
BioSequences.jl,Automa.jl,Kmers.jl
- Major contributions to
@kescobo- Senior Research Scientist at Wellesley College- Main bio packages
Microbiome.jlandBiobackeryUtils.jlmoved to EcoJulia 🤦
- Main bio packages
Other major early contributors
@jgreener64:BioStructures.jl@prcastro@kdm9
Packages and ecosystem
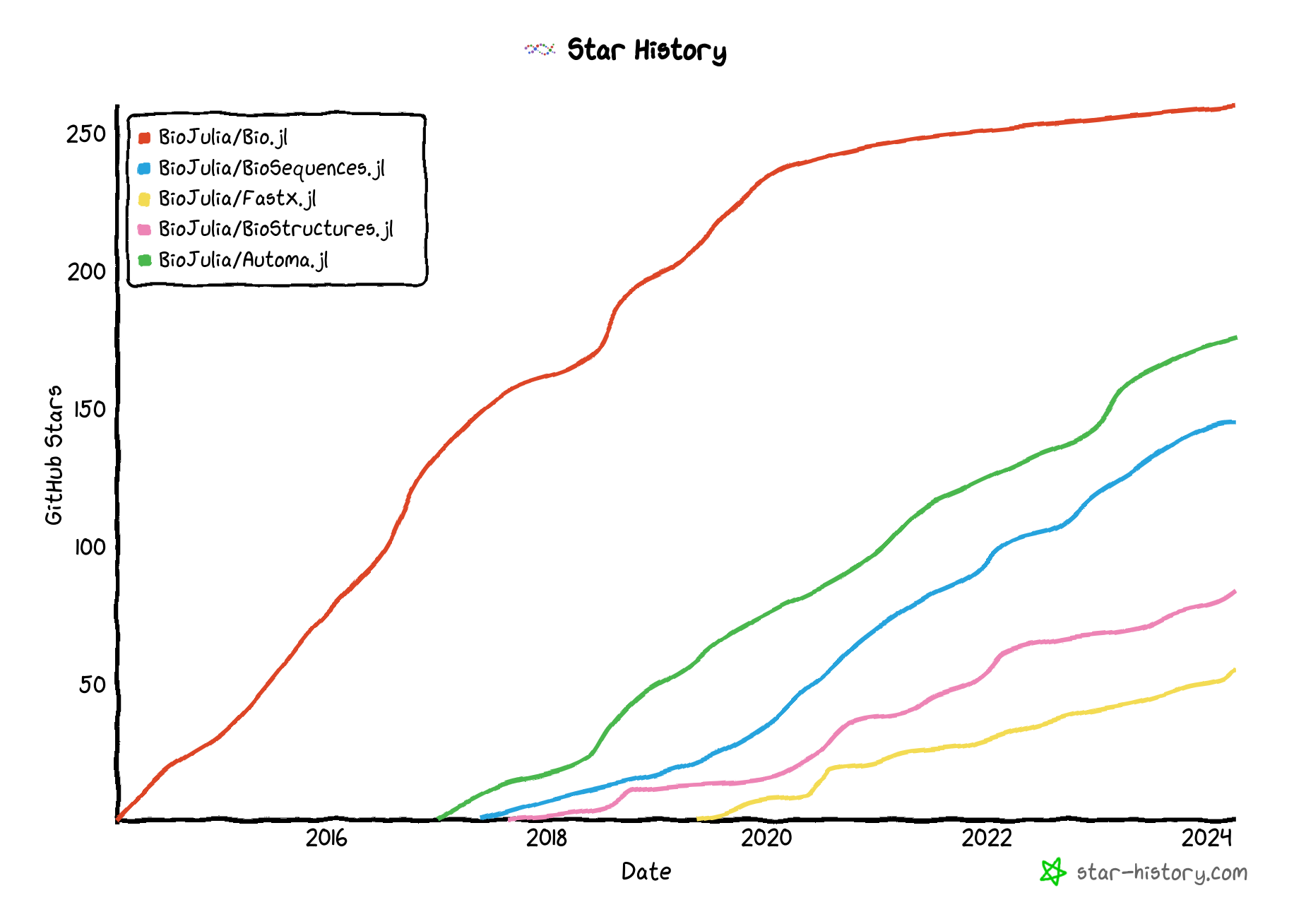
Non-BioJulia bio stuff in julia
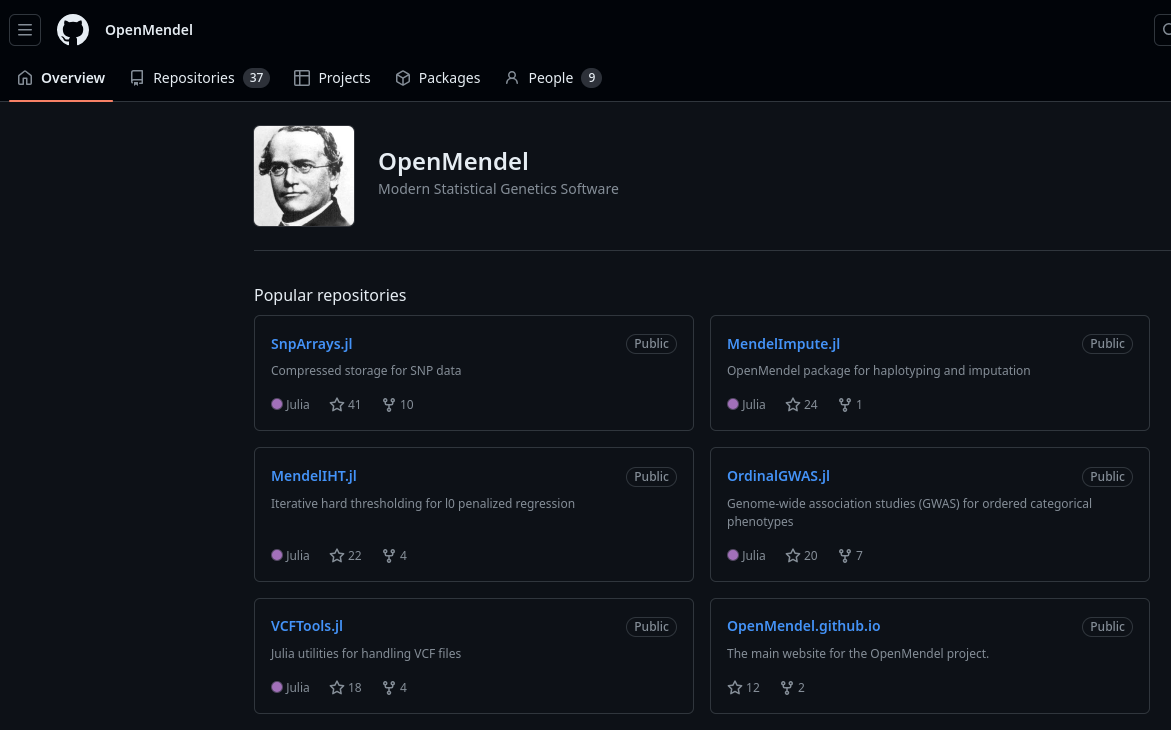
Non-BioJulia bio stuff in julia

Biology as fancy string manipulation
The central dogma of molecular biology
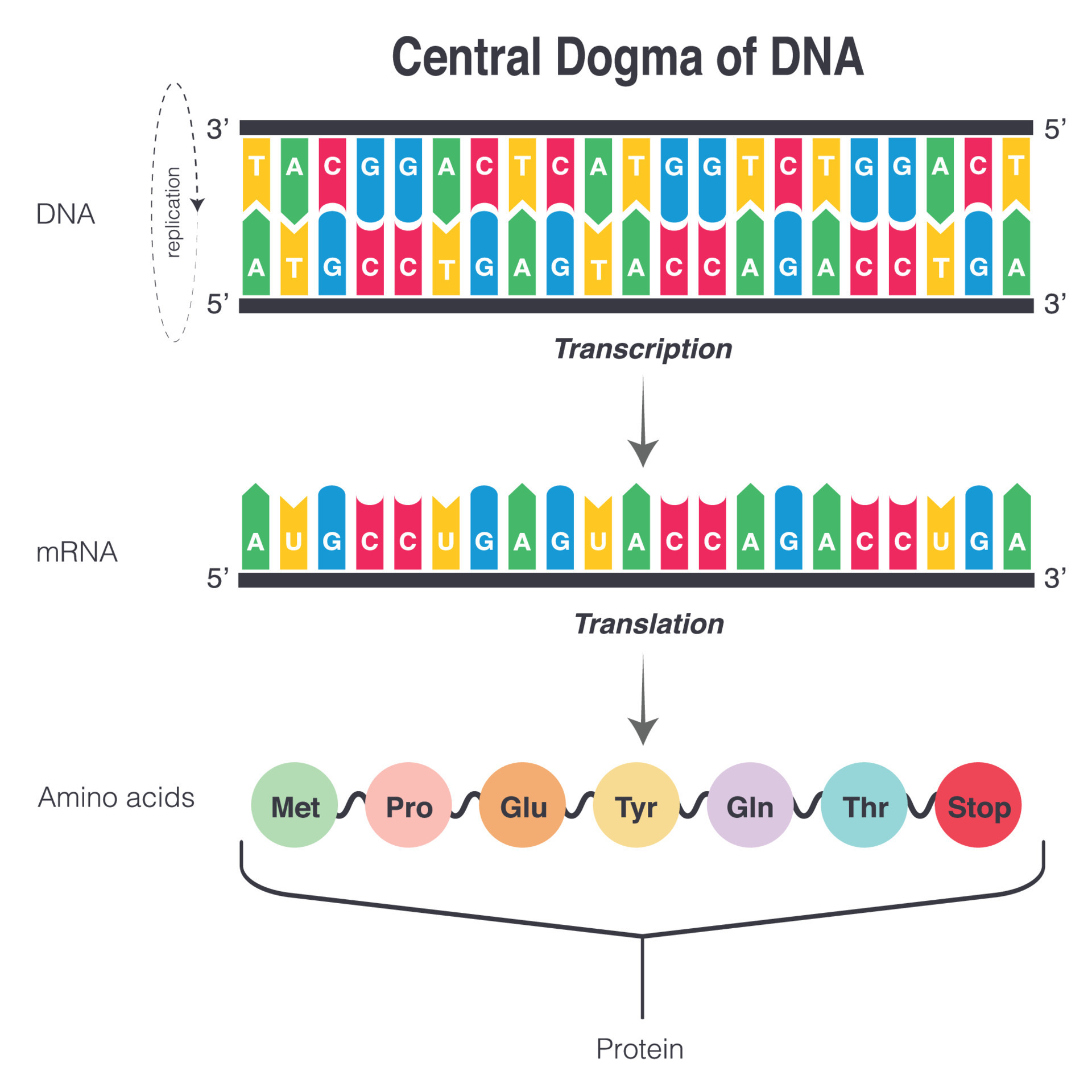
- DNA copied into DNA through “replication”
- DNA read into RNA through “transcription”
- RNA read into protein through “translation”
Major biological molecules are 1D arrays (polymers)
DNA
Protein
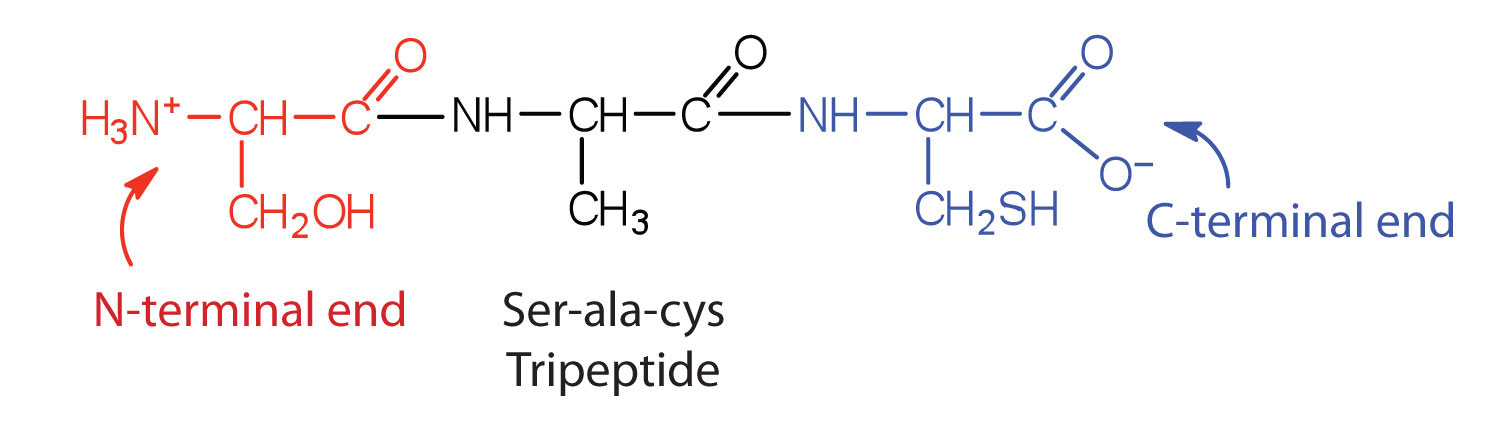
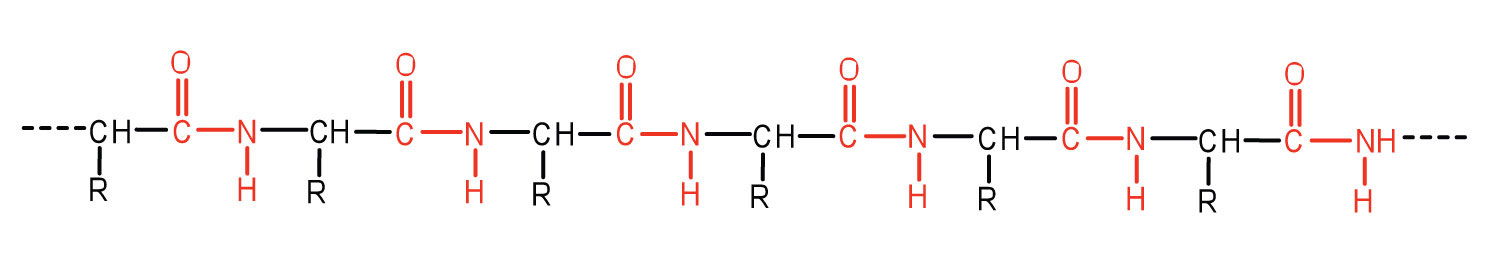
The functions of biomolecules are determined by their sequence
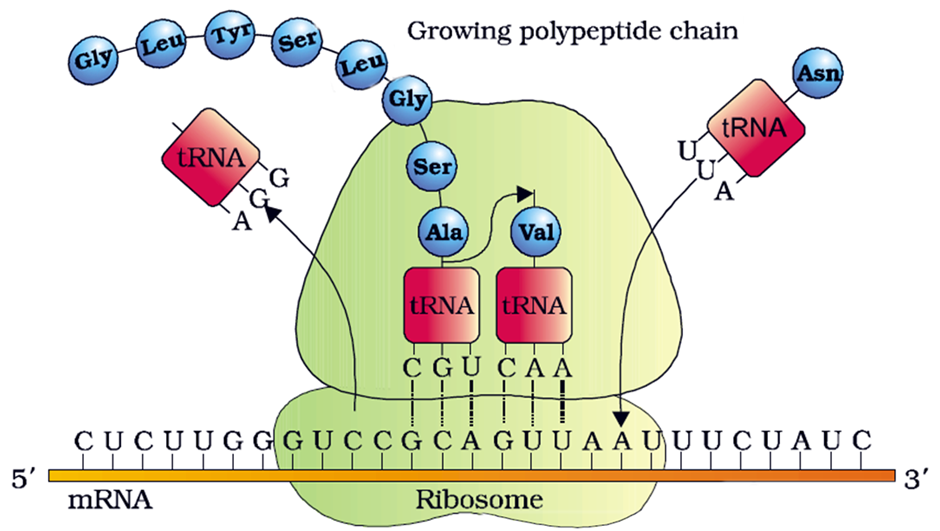
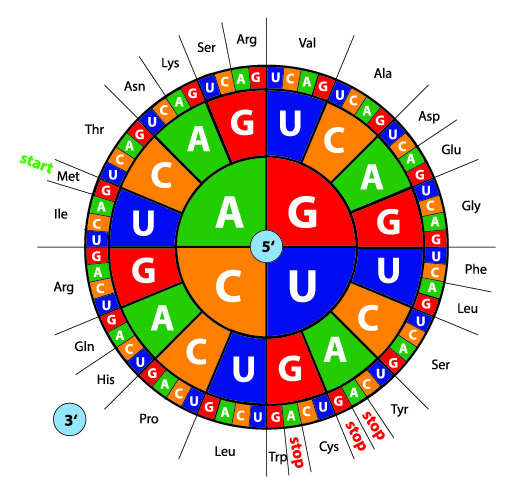
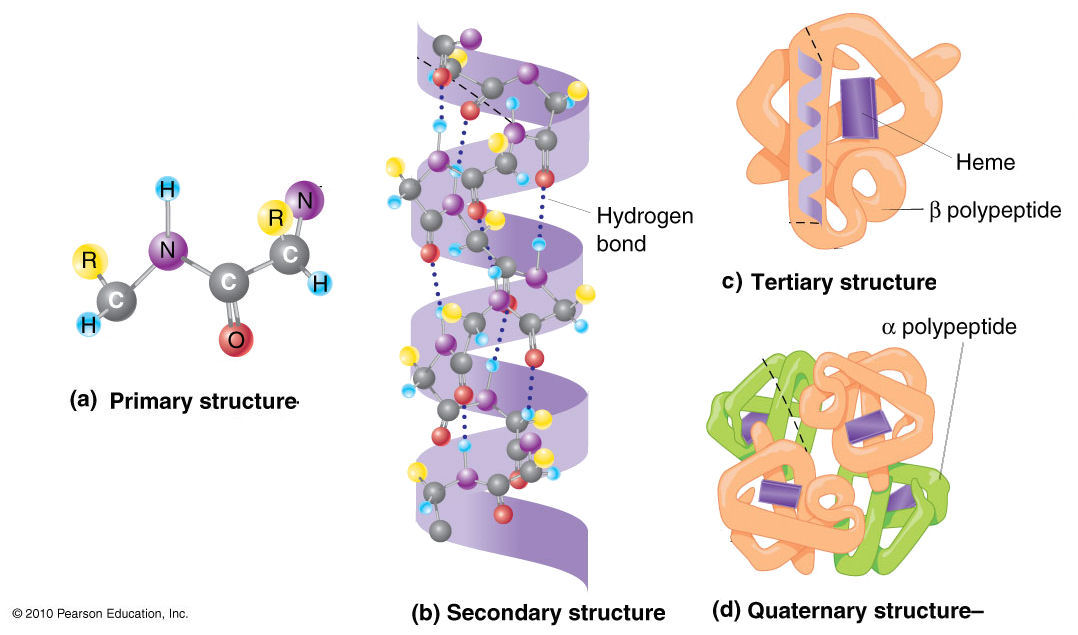
Evolutionary relationships are inferred from the relationship between sequences

Important biological algorithms are about alignment and search
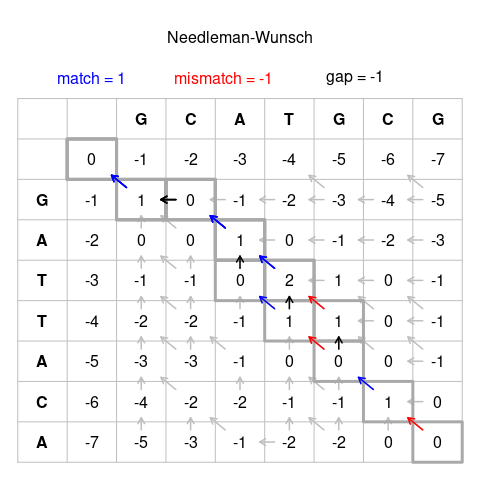
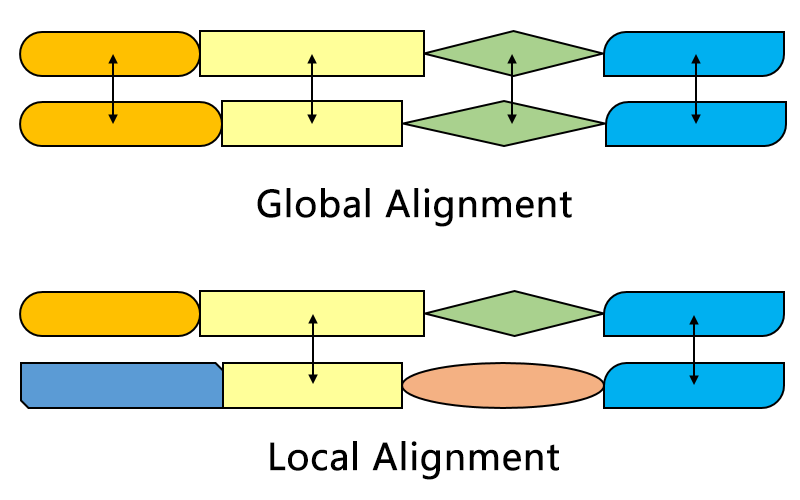
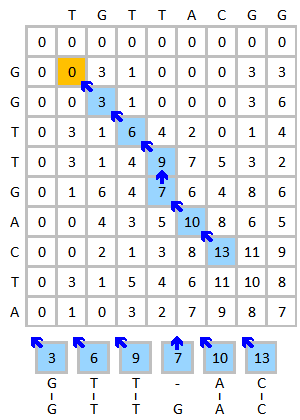
BioSequences.jl efficiently encodes biological sequences
import Base: summarysize
using BioSequences, Random
seq = randseq(DNAAlphabet{2}(), 512);
str = String(seq);
summarysize(seq) # 184
summarysize(str) # 520BioSequences.jl competes with special-purpose languages
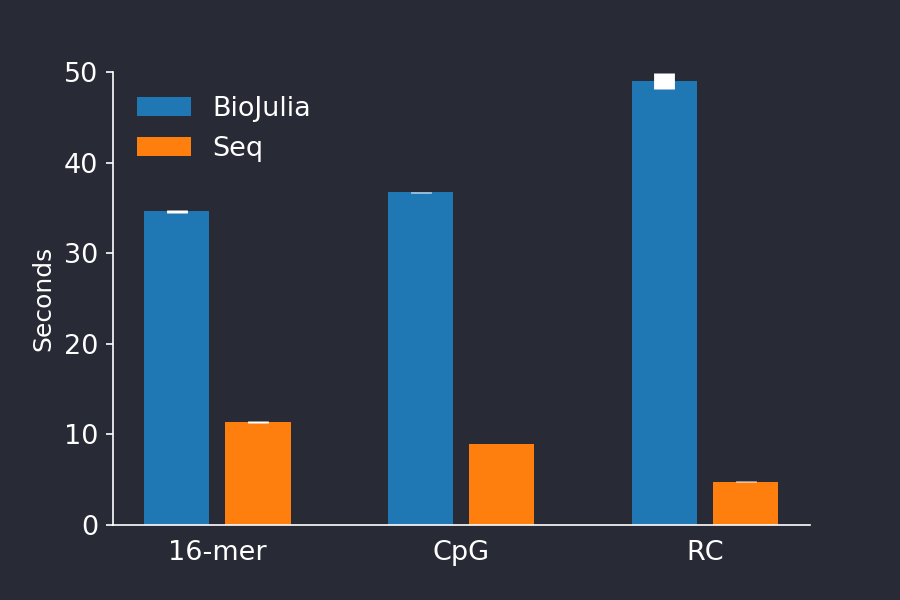
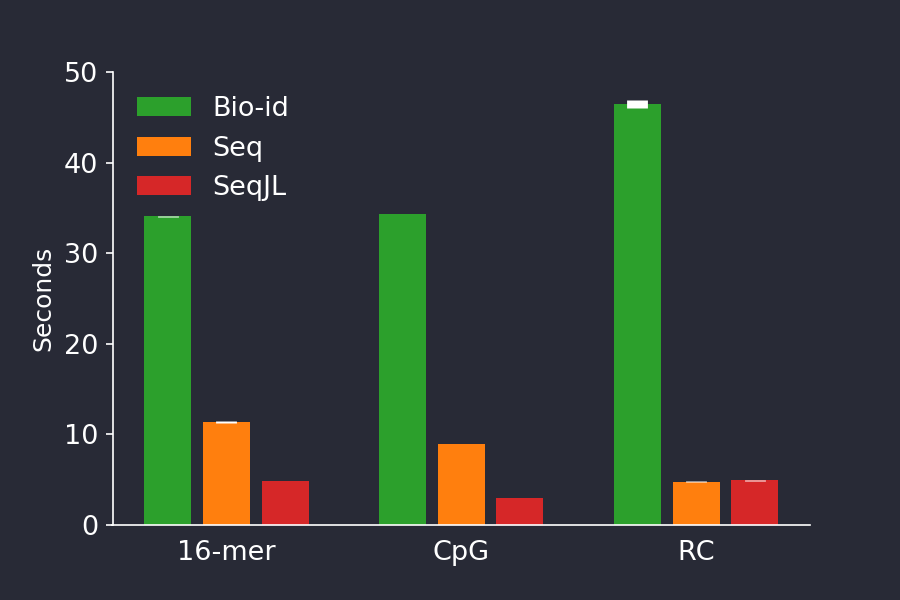
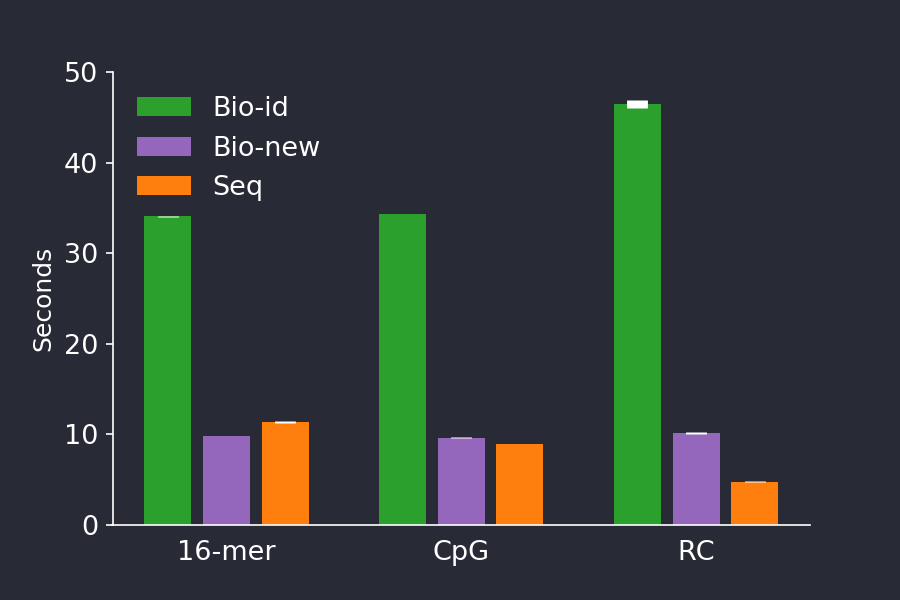
BioSequences.jl competes with special-purpose languages
File parsing is a critical part of bioinoformatics
FASTA
FASTQ
SAM
@HD VN:1.0 SO:unsorted
@SQ SN:1455__A0A0C2TZA5__A3781_04875 LN:1008
@SQ SN:1455__A0A0C2XRW0__A3781_18225 LN:804
@SQ SN:1455__A0A0C2XXQ4__A3781_14565 LN:867
...
VH01194:15:AAAWT2VHV:1:1101:49456:1398:N:0:GAACTGAGCG+CGCTCCACGA#0/1__1.101 16 1134687__A0A378ENW4__cobJ 67 3 150M * 0 0 CTGCAGGCGGCGGAAATCGTCGTCGGTTATAAAACTTACACCCATCTGGTGAAGGCTTTTACCGGCGACAAGCAGGTGATCAAAACCGGGATGTGCAAAGAGATTGAACGCTGTCAGGCGGCGATTGAACTGGCGCAGGCCGGGCACAAC CCCCCCCCCCCCCCCC;CCCCCCCCCCCC;CCCCCC;CCCCCCC;CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC AS:i:-59 XN:i:0 XM:i:12 XO:i:0 XG:i:0 NM:i:12 MD:Z:23T2C8C5G2C8A2C11T38G5C14G17T3 YT:Z:UU
VH01194:15:AAAWT2VHV:1:1101:31259:2098:N:0:GAACTGAGCG+CGCTCCACGA#0/1__1.441 16 73098__A0A1B7K6J8__M989_00754 328 40 150M * 0 0 CCAGCCGATTTCAGGAAATTAGGCCGTGATGCCGCGGCGACGCTGTTGTCGGTATCTAACGTAACGCTCTGGAATTCCATCGACTATTTCAGCCCCAGCGCCGAGCATAATCCTTTATTGATGACCTGGTCATTGGGCGTGGAAGAACAG CC-CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC;CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC;CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC;CCCCCCCCCCCCCC;CCCCCCCCCCCCCCCCC AS:i:-18 XN:i:0 XM:i:4 XO:i:0 XG:i:0 NM:i:4 MD:Z:2C23C18C16T87 YT:Z:UUThe development of every bioinformatics tool begins with the definition of a new file format, incompatible with all previous formats.
- Charles Darwin
Automa.jl enables the construction of correct but efficient file parsers
Automa makes Deterministic Finite Automata 
fasta_regex = let
header = re"[a-z]+"
seqline = re"[ACGT]+"
record = '>' * header * '\n' * rep1(seqline * '\n')
rep(record)
endmachine = let
header = onexit!(onenter!(re"[a-z]+", :mark_pos), :header)
seqline = onexit!(onenter!(re"[ACGT]+", :mark_pos), :seqline)
record = onexit!(re">" * header * '\n' * rep1(seqline * '\n'), :record)
compile(rep(record))
end

BioMakie.jl enables easy plotting of protein structure
using BioMakie
using GLMakie
using BioStructures
struc = retrievepdb("2vb1") |> Observable
## or
struc = read("2vb1.pdb", BioStructures.PDB) |> Observable
fig = Figure()
plotstruc!(fig, struc; plottype = :ballandstick, gridposition = (1,1), atomcolors = aquacolors)
plotstruc!(fig, struc; plottype = :covalent, gridposition = (1,2))
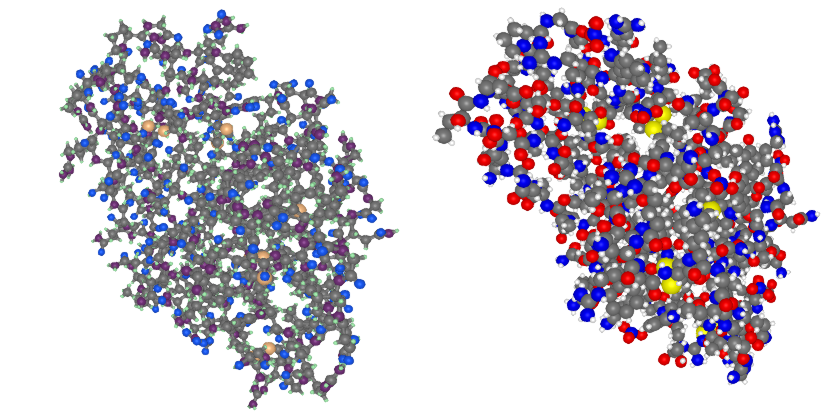
BioMakie.jl enables easy plotting of protein structure
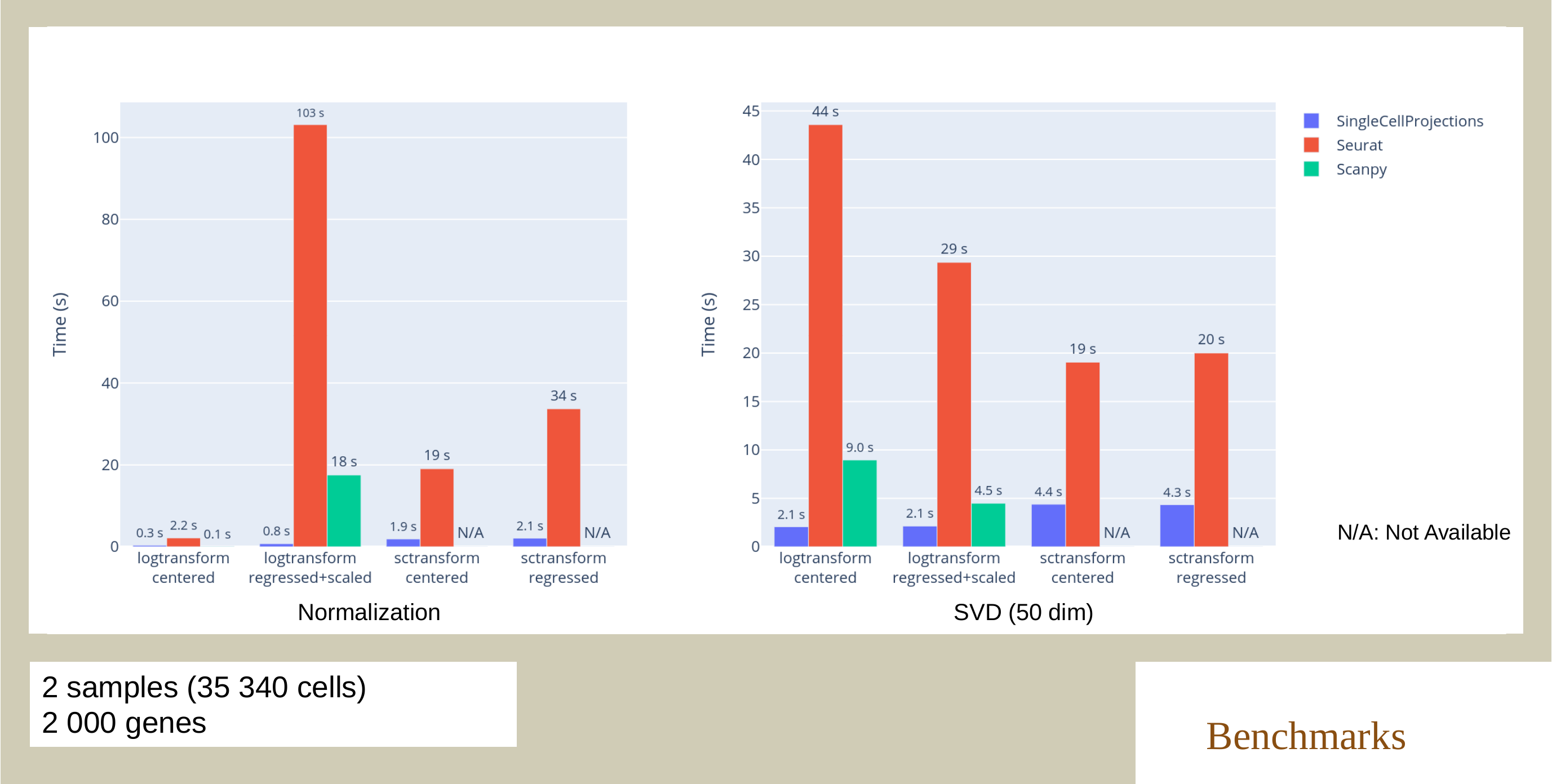
SingleCellProjections.jl enables fast, memory-efficient scRNAseq
SingleCellProjections.jl enables fast, memory-efficient scRNAseq
Biology as fancy data science
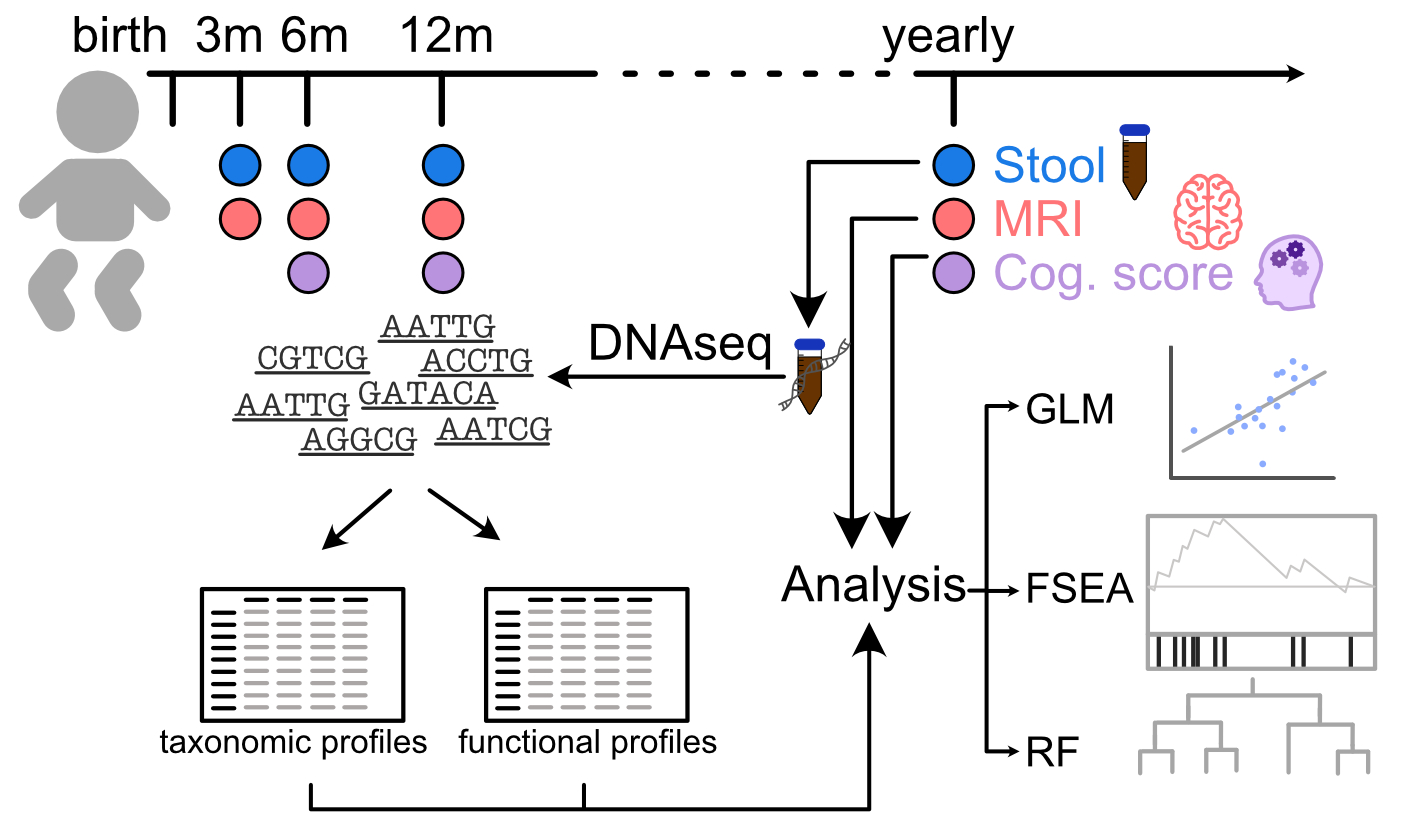
Pipeline for studying the gut microbiome
Microbiome.jl wraps sparse matrices
- Microbiome data is sparse
- species average ~5% prevalence
- genes average <1% prevalence
- Microbiome data is high-dimensional (~500 species, ~2M genes in 300 samples)
- Microbiome data has associated metadata
BiobakeryUtils.jl wraps command line tools

- Tools written in python with CLI
- Lots of python and binary dependencies (usually managed with
conda) - Each tool has large associated databases that are not installed along with the tool
BiobakeryUtils.jl wraps command line tools
$ humann --input some_reads.fastq.gz \
--taxonomic-profile some_profile.tsv --output ./ \
--threads 32 --remove-temp-output --search-mode uniref90 \
--output-basename somevs
julia> humann(; input="some_reads.fastq.gz",
taxonomic_profile="some_profile.tsv", output="./",
threads=32, remove_temp_output=true, search_mode="uniref90",
output_basename="some")Installation and deps managed by Conda.jl
Major gaps for bioinformatics adoption
Limitations of julia and wider ecosystem
- Many people doing computational biology aren’t computer scientists
- two-language problem doesn’t matter - they’re just using existing tools
- need lots of tutorials / examples (python and R have this covered)
- Most bioinformaticians run command line tools
- expect things to run from the command line, generate intermediate files
- Most bioinformaticians run reproducible (shell) pipelines
- eg Nextflow, snakemake
Limitations of BioJulia
- Not many tutorials / beginner guides
- Not many tools / wrappers that are easy to use
- Even fewer that are run from CLI
- Not many groups focus on julia for tool development or analysis
More resources
- JuliaCon Talks
- SciML SysBio: https://live.juliacon.org/talk/GQPUWV
SingleCellProjections.jl: https://live.juliacon.org/talk/NPADF7BioMakie.jl: https://www.youtube.com/watch?v=-C7Zbh6UTgk
- Lots of blog posts by Jakob Nybo Nissen https://viralinstruction.com
- Discourse: https://discourse.julialang.org/c/domain/bio/
Thanks!
Questions?